BFM in a few more words
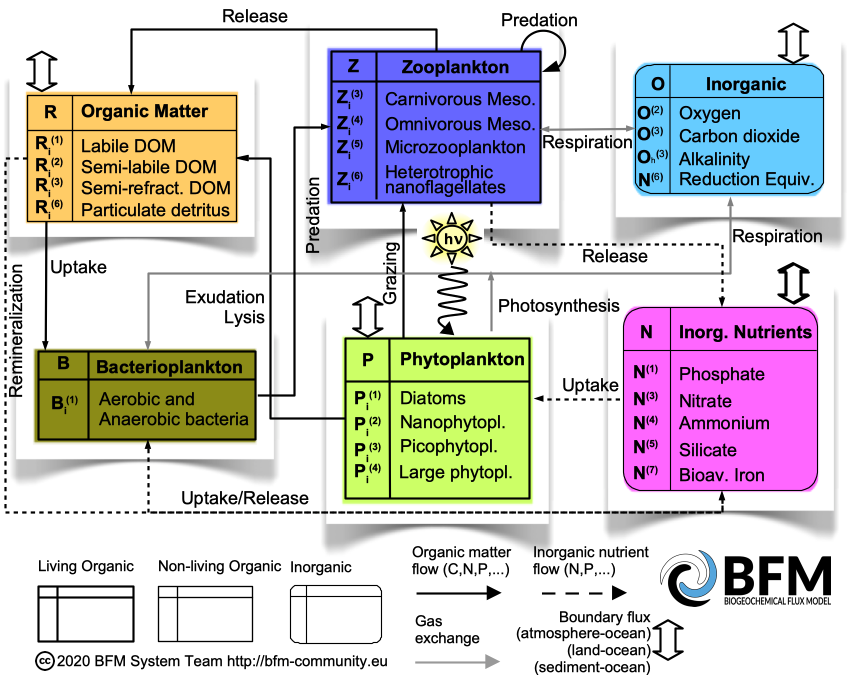
The Biogeochemical Flux Model is a biomass-based numerical model to simulate the major biogeochemical processes occurring in pelagic, benthic and sea-ice components of marine ecosystems. The BFM considers the cycles of nitrogen, phosphorus, silica, iron, carbon, and oxygen in the water, sediments, sea ice and in the related organisms from bacteria to benthic meiofauna. All living organisms are Plankton dynamics are parameterized in terms of functional groups with prescribed functional traits. For instance, the BFM plankton functional groups are subdivided in producers (phytoplankton), consumers (zooplankton), and decomposers (bacteria). These broad functional classifications are further partitioned into functional subgroups to create a planktonic food web (e.g. diatoms, picophytoplankton, microzooplankton, etc.).
The BFM structure is flexible and modular, so that the number, type and properties of the functional groups, as well as their chemical constituents, can be increased or modified. Different model configurations can be used to focus on different aspects. The code is open source and written in the FORTRAN90 language. It is a scientific, non-commercial software developed to be compliant with unix/linux environments.
The BFM model can be used in standalone mode to study closed systems like laboratory cultures or coupled to numerical hydrodynamic models to include transport and spatial heterogeneity. It has been successfully coupled to the Princeton Ocean Model (POM), OGS Transport Model (OGSTM), MIT General Circulation Model (MIT-GCM), FABM programming framework (FABM), and Nucleus for European Modelling of the Ocean (NEMO) and applied at the global ocean, regional, subregional and local space scales. It has been used for a variety of applications from forecasting to global climate simulations (check the scientific bibliography).
Contact the BFM system team if you are interested in couplings with other numerical ocean models.